Classification Methods
1 Introduction to Classification Methods
When we apply cluster analysis to a dataset, we let the values of the variables
that were measured tell us if there is any structure to the observations in the
data set, by choosing a suitable metric and seeing if groups of observations that
are all close together can be found. If we have an auxilliary variable (like the
country of origin from the cars example), it may be interesting to see if the
natural clustering of the data corresponds to this variable, but it's important to
remember that the idea of clustering is just to see if any groups form naturally,
not to see if we can actually figure out which group an observation belongs to
based on the values of the variables that we have.
When the true goal of our data analysis is to be able to predict which of several
non-overlapping groups an observation belongs to, the techniques we use are known
as classification techniques. We'll take a look at three classification techniques:
kth nearest neighbor classification, linear discrimininant analysis, and recursive
partitioning.
2 kth Nearest Neighbor Classification
The idea behind nearest neighbor classification is simple and somewhat intuitive -
find other observations in the data that are close to an observation we're interested,
and classify that observation based on the class of its neighbors. The number of
neighbors that we consider is where the "k" comes in - usually we'll have to look
at several different values of k to determine which ones work well with a particular
data set. Values in the range of one to ten are usually reasonable choices.
Since we need to look at all of the distances between one observation and all the
others in order to find the neighbors, it makes sense to form a distance matrix before
starting a nearest neighbor classification. Each row of the distance matrix tells
us the distances to all the other observations, so we need to find the k smallest
values in each row of the distance matrix. Once we find those smallest values, we
determine which observations they belong to, and look at how those observations were
classified. We assign whichever value of the classification that was most common among
the k nearest neighbors as our guess (predicted value) for the current observation, and
then move on to
the next row of the distance matrix. Once we've looked at every row of the distance
matrix, we'll have classified every observation, and can compare the predicted
classification with the actual classification. To see how well we've done, various
error rates can be examined. If the observations are classified as TRUE / FALSE,
for example disease or no disease, then we can look at two types of error rates.
The first type of error rate, known as Type I error, occurs when we say that an
observation should be classified as TRUE when it really should have been FALSE. The
other type of error (Type II) occurs when we say that an observation should be
classified as FALSE when it should have been TRUE. When the classification is something
other than TRUE/FALSE, we can report an overall error rate, that is, the fraction of
observations for which our prediction was not correct. In either case, the error
rates can be calculated in R by using the table function. As a simple
example, suppose we have two vectors: actualvalues, which contains the actual values
of a classification variable, and predvalues, the value that our classification
predicted:
> actualvalues = c(TRUE,TRUE,TRUE,FALSE,FALSE,TRUE,FALSE,TRUE,FALSE,FALSE)
> predvalues = c(TRUE,TRUE,TRUE,TRUE,FALSE,FALSE,FALSE,TRUE,FALSE,TRUE)
> tt = table(actualvalues,predvalues)
> tt
predvalues
actualvalues FALSE TRUE
FALSE 3 2
TRUE 1 4
The observations that contribute to Type I error (the actual value is false but we
predicted true) can be found in the first row and second column; those that
contribute to Type II error can be found in the second row and first column. Since
the table function returns a matrix, we can calculate the rows as
follows:
> tot = sum(tt)
> type1 = tt['FALSE','TRUE'] / tot
> type2 = tt['TRUE','FALSE'] / tot
> type1
[1] 0.2
> type2
[1] 0.1
3 Cross Validation
There's one problem with the above scheme. We used the data that we're making
predictions about in the process of making those predictions.
In other words, the data that we're making predictions for is not independent of
the data that we're using to make the predictions. As might be expected, it's been
shown in practice that calculating error rates this way will almost always make our
classification method look better than it should be. If the data can be naturally
(or even artificially) divided into two groups, then one can be used as a training
set, and the other can be used as a test set - we'd calculate our error rates only
from the classification of the test set using the training set to make our predictions.
Many statisticians don't like the idea of having to "hold back" some of their data
when building models, so an alternative way to bring some independence to our predictions
known as v-fold cross validation has been devised. The idea is to first divide the entire
data set
into v groups. To classify objects in the first group, we don't use any of the first
group to make our predictions; in the case of k-th nearest neighbor classification,
that would mean that when we're looking for the smallest distances in order to classify
an observation, we don't consider any of the distances corresponding to other members
of the same group that the current one belongs to. The basic idea is that we want
to make the prediction for an observation as independent from that observation as we
can. We continue through each of the v groups, classifying observations in each group
using only observations from the other groups. When we're done we'll have a prediction
for each observation, and can compare them to the actual values as in the previous
example.
Another example of cross-validation is leave-out-one cross-validation. With this method,
we predict the classification of an observation without using the observation itself.
In other words, for each observation, we perform the analysis without using that
observation, and then predict where that observation would be classified using that
analysis.
4 Linear Discriminant Analysis
One of the oldest forms of classification is known as linear discriminant analysis.
The idea is to form linear combinations of predictor variables (similar to a linear
regression model) in such a way that the average value of these linear combinations will
be as different as possible for the different levels of the classification variable.
Based on the values of the linear combinations, linear discriminant analysis reports
a set of posterior probabilities for every level of the classification, for each
observation, along with the level of the classification variable that the analysis
predicted. Suppose we have a classification variable that can take one of three
values: after a linear discriminant analysis, we will have three probabilities (adding
up to one) for each variable that tell how likely it is that the observation
be categorized into each of the three categories; the predicted classificiation is the
one that had the highest probability, and we can get insight into the quality of the classification
by looking at the values of the probabilities.
To study the different classification methods, we'll use a data set about
different wines. This data set contains various measures regarding chemical
and other properties of the wines, along with a variable identifying the
Cultivar (the particular variety of the grape from which the wine was produced).
We'll try to classify the observations based on the Cultivar, using the other
variables.
The data is available at http://www.stat.berkeley.edu/~spector/s133/data/wine.data; information about
the variables is at http://www.stat.berkeley.edu/~spector/s133/data/wine.names
First, we'll read in the wine dataset:
wine = read.csv('http://www.stat.berkeley.edu/~spector/s133/data/wine.data',header=FALSE)
names(wine) = c("Cultivar", "Alcohol", "Malic.acid", "Ash", "Alkalinity.ash",
"Magnesium", "Phenols", "Flavanoids", "NF.phenols", "Proanthocyanins",
"Color.intensity","Hue","OD.Ratio","Proline")
wine$Cultivar = factor(wine$Cultivar)
Notice that I set wine$Cultivar to be a factor. Factors are very
important and useful in modeling functions because categorical variables almost always
have to be treated differently than numeric variables, and turning a categorical variable
into
a factor will insure that they are always used properly in modeling functions. Not
surprisingly, the dependent variable for lda must be a factor.
The class library of R provides two functions for nearest neighbor
classification. The first, knn, takes the approach of using a
training set and a test set, so it would require holding back some of the
data. The other function, knn.cv uses leave-out-one cross-validation,
so it's more suitable to use on an entire data set.
Let's use knn.cv on the wine data set. Since, like cluster analysis,
this technique is based
on distances, the same considerations regarding standardization as we saw
with cluster analysis apply. Let's examine a summary for the data frame:
> summary(wine)
Cultivar Alcohol Malic.acid Ash Alkalinity.ash
1:59 Min. :11.03 Min. :0.740 Min. :1.360 Min. :10.60
2:71 1st Qu.:12.36 1st Qu.:1.603 1st Qu.:2.210 1st Qu.:17.20
3:48 Median :13.05 Median :1.865 Median :2.360 Median :19.50
Mean :13.00 Mean :2.336 Mean :2.367 Mean :19.49
3rd Qu.:13.68 3rd Qu.:3.083 3rd Qu.:2.558 3rd Qu.:21.50
Max. :14.83 Max. :5.800 Max. :3.230 Max. :30.00
Magnesium Phenols Flavanoids NF.phenols
Min. : 70.00 Min. :0.980 Min. :0.340 Min. :0.1300
1st Qu.: 88.00 1st Qu.:1.742 1st Qu.:1.205 1st Qu.:0.2700
Median : 98.00 Median :2.355 Median :2.135 Median :0.3400
Mean : 99.74 Mean :2.295 Mean :2.029 Mean :0.3619
3rd Qu.:107.00 3rd Qu.:2.800 3rd Qu.:2.875 3rd Qu.:0.4375
Max. :162.00 Max. :3.880 Max. :5.080 Max. :0.6600
Proanthocyanins Color.intensity Hue OD.Ratio
Min. :0.410 Min. : 1.280 Min. :0.4800 Min. :1.270
1st Qu.:1.250 1st Qu.: 3.220 1st Qu.:0.7825 1st Qu.:1.938
Median :1.555 Median : 4.690 Median :0.9650 Median :2.780
Mean :1.591 Mean : 5.058 Mean :0.9574 Mean :2.612
3rd Qu.:1.950 3rd Qu.: 6.200 3rd Qu.:1.1200 3rd Qu.:3.170
Max. :3.580 Max. :13.000 Max. :1.7100 Max. :4.000
Proline
Min. : 278.0
1st Qu.: 500.5
Median : 673.5
Mean : 746.9
3rd Qu.: 985.0
Max. :1680.0
Since the scale of the variables differ widely, standardization
is probably a good idea. We'll divide each variable by its standard deviation
to try to give each variable more equal weight in determining the distances:
> wine.use = scale(wine[,-1],scale=apply(wine[,-1],2,sd))
> library(class)
> res = knn.cv(wine.use,wine$Cultivar,k=3)
> names(res)
NULL
> length(res)
[1] 178
Since there are no names, and the length of res is the same as the
number of observations, knn.cv is simply returning the classifications
that the method predicted for each observation using leave-out-one cross validation.
This means we can compare the predicted values to the true values using table:
> table(res,wine$Cultivar)
res 1 2 3
1 59 4 0
2 0 63 0
3 0 4 48
To calculate the proportion of incorrect classifications, we can use the row
and col functions. These unusual functions don't seem to do anything very
useful when we simply call them:
> tt = table(res,wine$Cultivar)
> row(tt)
[,1] [,2] [,3]
[1,] 1 1 1
[2,] 2 2 2
[3,] 3 3 3
> col(tt)
[,1] [,2] [,3]
[1,] 1 2 3
[2,] 1 2 3
[3,] 1 2 3
However, if you recall that the misclassified observations are
those that are off the diagonal, we can find those observations as follows:
> tt[row(tt) != col(tt)]
[1] 0 0 4 4 0 0
and the proportion of misclassified observations can be calculated
as:
> sum(tt[row(tt) != col(tt)]) / sum(tt)
[1] 0.04494382
or a missclassification rate of about 4.5
Could we have done better if we used 5 nearest neighbors instead of 3?
> res = knn.cv(wine.use,wine$Cultivar,k=5)
> tt = table(res,wine$Cultivar)
> sum(tt[row(tt) != col(tt)]) / sum(tt)
[1] 0.02808989
How about using just the single nearest neighbor?
> res = knn.cv(wine.use,wine$Cultivar,k=1)
> tt = table(res,wine$Cultivar)
> sum(tt[row(tt) != col(tt)]) / sum(tt)
[1] 0.04494382
For this data set, using k=5 did slightly better than 1 or 3.
In R, linear discriminant analysis is provided by the lda function from the
MASS library, which is part of the base R distribution.
Like many modeling and analysis functions in R, lda takes a formula as its
first argument. A formula in R is a way of describing a set of relationships that
are being studied. The dependent variable, or the variable to be predicted, is
put on the left hand side of a tilda (~) and the variables that will be used
to model or predict it are placed on the right hand side of the tilda, joined together
by plus signs (+). To save typing, you an provide the name of a data frame
through the data= argument, and use the name of the variables in the data
frame in your formula without retyping the data frame name or using the with
function.
A convenience offered by the modeling functions is that a period (.)
on the right-hand side of the tilda in a formula is interpreted as meaning "all the
other variables in the data frame, except the dependent variable". So a very popular
method of specifying a formula is to use the period, and then use subscripting to
limit the data= argument to just the variables you want to fit. In this
example, we don't need to do that, because we really do want to use all the variables
in the data set.
> wine.lda = lda(Cultivar ~ .,data=wine)
We'll see that most of the modeling functions in R share many things in common. For
example, to predict values based on a model, we pass the model object to the
predict function along with a data frame containing the observations
for which we want predictions:
> pred = predict(wine.lda,wine)
To see what's available from the call to predict, we can look at the names of the
pred object:
> names(pred)
[1] "class" "posterior" "x"
The predicted classification is stored in the class component of the
object returned by predict
Now that we've got the predicted classes, we can see how well the classification
went by making a cross-tabulation of the real Cultivar with our prediction, using
the table function:
> table(wine$Cultivar,pred$class)
predclass
1 2 3
1 59 0 0
2 0 71 0
3 0 0 48
Before we get too excited about these results, remember the caution about predicting
values based on models that were built using those values. The error rate we see in
the table (0) is probably an overestimate of how good the classification rule is.
We can use v-fold cross validation on the data, by using the lda command
repeatedly to classify groups of observations (folds) using the rest of the data
to build the model.
We could write this out "by hand", but it would be useful
to have a function that could do this for us. Here's such a function:
vlda = function(v,formula,data,cl){
require(MASS)
grps = cut(1:nrow(data),v,labels=FALSE)[sample(1:nrow(data))]
pred = lapply(1:v,function(i,formula,data){
omit = which(grps == i)
z = lda(formula,data=data[-omit,])
predict(z,data[omit,])
},formula,data)
wh = unlist(lapply(pred,function(pp)pp$class))
table(wh,cl[order(grps)])
}
This function accepts four arguments: v, the number of folds in the cross
classification, formula which is the formula used in the linear discriminant
analysis, data which is the data frame to use, and cl,
the classification variable (wine$Cultivar in this case).
By using the sample function, we make sure that the groups that are used for
cross-validation aren't influenced by the ordering of the data - notice how the
classification variable (cl) is indexed by order(grps) to make sure
that the predicted and actual values line up properly.
Applying this function to the wine data will give us a better idea of the
actual error rate of the classifier:
> vlda(5,Cultivar~.,wine,wine$Cultivar)
wh 1 2 3
1 59 1 0
2 0 69 1
3 0 1 47
While the error rate is still very good, it's not quite perfect:
> error = sum(tt[row(tt) != col(tt)]) / sum(tt)
> error
[1] 0.01685393
Note that because of the way we randomly divide the observations, you'll see
a slightly different table every time you run the vlda function.
We could use a similar method to apply v-fold cross-validation to the kth
nearest neighbor classification. Since the knn function accepts a
training set and a test set, we can make each fold a test set, using the
remainder of the data as a training set. Here's a function to apply this
idea:
vknn = function(v,data,cl,k){
grps = cut(1:nrow(data),v,labels=FALSE)[sample(1:nrow(data))]
pred = lapply(1:v,function(i,data,cl,k){
omit = which(grps == i)
pcl = knn(data[-omit,],data[omit,],cl[-omit],k=k)
},data,cl,k)
wh = unlist(pred)
table(wh,cl[order(grps)])
}
Let's apply the function to the standardized wine data:
> tt = vknn(5,wine.use,wine$Cultivar,5)
> tt
wh 1 2 3
1 59 2 0
2 0 66 0
3 0 3 48
> sum(tt[row(tt) != col(tt)]) / sum(tt)
[1] 0.02808989
Note that this is the same misclassification rate as
acheived by the "leave-out-one" cross validation provided by
knn.cv.
Both the nearest neighbor and linear discriminant methods make it
possible to classify new observations, but they don't give much
insight into what variables are important in the classification.
The scaling element of the object returned by lda
shows the linear combinations of the original variables that were created
to distinguish between the groups:
> wine.lda$scaling
LD1 LD2
Alcohol -0.403399781 0.8717930699
Malic.acid 0.165254596 0.3053797325
Ash -0.369075256 2.3458497486
Alkalinity.ash 0.154797889 -0.1463807654
Magnesium -0.002163496 -0.0004627565
Phenols 0.618052068 -0.0322128171
Flavanoids -1.661191235 -0.4919980543
NF.phenols -1.495818440 -1.6309537953
Proanthocyanins 0.134092628 -0.3070875776
Color.intensity 0.355055710 0.2532306865
Hue -0.818036073 -1.5156344987
OD.Ratio -1.157559376 0.0511839665
Proline -0.002691206 0.0028529846
It's really not that easy to interpret them, but variables with
large absolute values in the scalings are more likely to influence the
process. For this data, Flavanoids, NF.phenols, Ash,
and Hue seem to be among the important variables.
A different way to get some insight into this would be to examine the
means of each of the variables broken down by the classification
variable. Variables which show a large difference among the
groups would most likely be the ones that are useful in predicting
which group an observation belongs in. One graphical way of
displaying this information is with a barplot. To make sure we
can see differences for all the variables, we'll use the
standardized version of the data:
> mns = aggregate(wine.use,wine['Cultivar'],mean)
> rownames(mns) = mns$Cultivar
> mns$Cultivar = NULL
> barplot(as.matrix(mns),beside=TRUE,cex.names=.8,las=2)
The las parameter rotates the labels on
the x-axis so that we can see them all. Here's the plot:
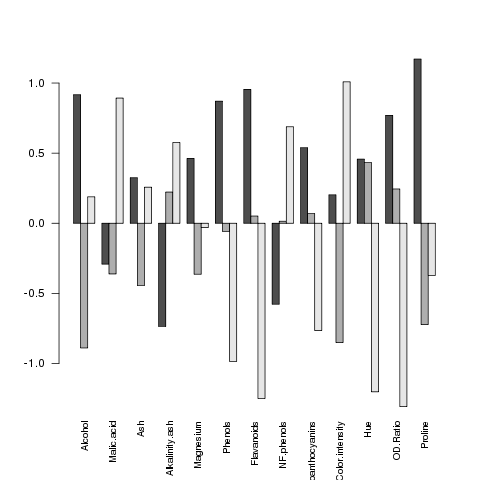 It seems like the big differences are found in Flavanoids,
Hue and OD.Ratio
Now let's take a look at a method that makes it very clear which variables
are useful in distinguishing among the groups.
It seems like the big differences are found in Flavanoids,
Hue and OD.Ratio
Now let's take a look at a method that makes it very clear which variables
are useful in distinguishing among the groups.
5 Recursive Partitioning
An alternative classification method, developed in the 1980s, has attracted a
lot of attention in a variety of different fields. The technique, known as
recursive partitioning or CART (Classification and Regression Trees), can be
use for either classification or regression - here we'll concentrate on its
use for classification. The basic idea is to examine, for each of the variables
that we're using in our classification model, the results of splitting the data
set based on one of the values of that variable, and then examining how that
split helps us distinguish between the different classes of interest. For example,
suppose we're using just one variable to help distinguish between two classes:
> mydata = data.frame(x=c(7,12,8,3,4,9,2,19),grp=c(1,2,1,1,1,2,2,2))
> mydata
x grp
1 7 1
2 12 2
3 8 1
4 3 1
5 4 1
6 9 2
7 2 2
8 19 2
We'd consider each value of x in the data, and split the data into two
groups: one where x was less than or equal to the value and the other where
x
is greater than the value. We then look at how our classification variable (grp
in this example) breaks down when the data is split this way. For this example, we
can look at all the possible cross-tabulations:
> tbls = sapply(mydata$x,function(val)table(mydata$x <= val,mydata$grp))
> names(tbls) = mydata$x
> tbls
> tbls
$"7"
1 2
FALSE 1 3
TRUE 3 1
$"12"
1 2
FALSE 0 1
TRUE 4 3
$"8"
1 2
FALSE 0 3
TRUE 4 1
$"3"
1 2
FALSE 3 3
TRUE 1 1
$"4"
1 2
FALSE 2 3
TRUE 2 1
$"9"
1 2
FALSE 0 2
TRUE 4 2
$"2"
1 2
FALSE 4 3
TRUE 0 1
$"19"
1 2
TRUE 4 4
If you look at each of these tables, you can see that when we split with the rule
x <= 8, we get the best separation; all three of the cases where
x is greater than
8 are classified as 2, and four out of five of the cases where x is less than
8 are classified as 1.
For this phase of a recursive partitioning, the rule x <= 8 would be
chosen to split the data.
In real life situations, where there will always be more than one variable, the
process would be repeated for each of the variables, and the chosen split would
the one from among all the variables that resulted in the best separation or
node purity as it is sometimes known. Now that we've found the single best split,
our data is divided into two groups based on that split. Here's where the recursive
part comes in: we keep doing the same thing to each side of the split data, searching
through all values of all variables until we find the one that gives us the best
separation, splitting the data by that value and then continuing. As implemented in
R through the rpart function in the rpart library, cross validation
is used internally to determine when we should stop splitting the data, and present a
final tree as the output. There are also options to the rpart function specifying the
minimum number of observations allowed in a split, the minimum number of observations
allowed in one of the final nodes and the maximum number of splits allowed, which can
be set through the control= argument to rpart. See the help page
for rpart.control for more information.
To show how information from recursive partitioning is displayed, we'll use the same data
that we used with lda. Here are the commands to run the analysis:
> library(rpart)
> wine.rpart = rpart(Cultivar~.,data=wine)
> wine.rpart
n= 178
node), split, n, loss, yval, (yprob)
* denotes terminal node
1) root 178 107 2 (0.33146067 0.39887640 0.26966292)
2) Proline>=755 67 10 1 (0.85074627 0.05970149 0.08955224)
4) Flavanoids>=2.165 59 2 1 (0.96610169 0.03389831 0.00000000) *
5) Flavanoids< 2.165 8 2 3 (0.00000000 0.25000000 0.75000000) *
3) Proline< 755 111 44 2 (0.01801802 0.60360360 0.37837838)
6) OD.Ratio>=2.115 65 4 2 (0.03076923 0.93846154 0.03076923) *
7) OD.Ratio< 2.115 46 6 3 (0.00000000 0.13043478 0.86956522)
14) Hue>=0.9 7 2 2 (0.00000000 0.71428571 0.28571429) *
15) Hue< 0.9 39 1 3 (0.00000000 0.02564103 0.97435897) *
The display always starts at the root (all of the data), and reports the splits in the order
they occured. So after examining all possible values of all the variables, rpart
found that the Proline variable did the best job of dividing up the observations into
the different cultivars; in particular of the 67 observations for which Proline
was greater than 755, the fraction that had Cultivar == 1 was .8507. Since there's
no asterisk after that line, it indicates that rpart could do a better job by considering
other variables. In particular if Proline was >= 755, and Flavanoids
was >= 2.165, the fraction of Cultivar 1 increases to .9666; the asterisk at the end of a line
indicates that this is a terminal node, and, when classifying an observation if Proline
was >= 755 and Flavanoids was >= 2.165, rpart would immediately assign it to
Cultivar 1 without having to consider any other variables. The rest of the output can be determined
in a similar fashion.
An alternative way of viewing the output from rpart is a tree diagram, available through
the plot function. In order to identify the parts of the diagram, the text function
also needs to be called.
> plot(wine.rpart)
> text(wine.rpart,use.n=TRUE,xpd=TRUE)
The xpd=TRUE is a graphics parameter that is useful when a plot gets truncated, as sometimes
happens with rpart plots. There are other options to plot and text which
will change the appearance of the output; you can find out more by looking at the help pages for
plot.rpart and text.rpart. The graph appears below.
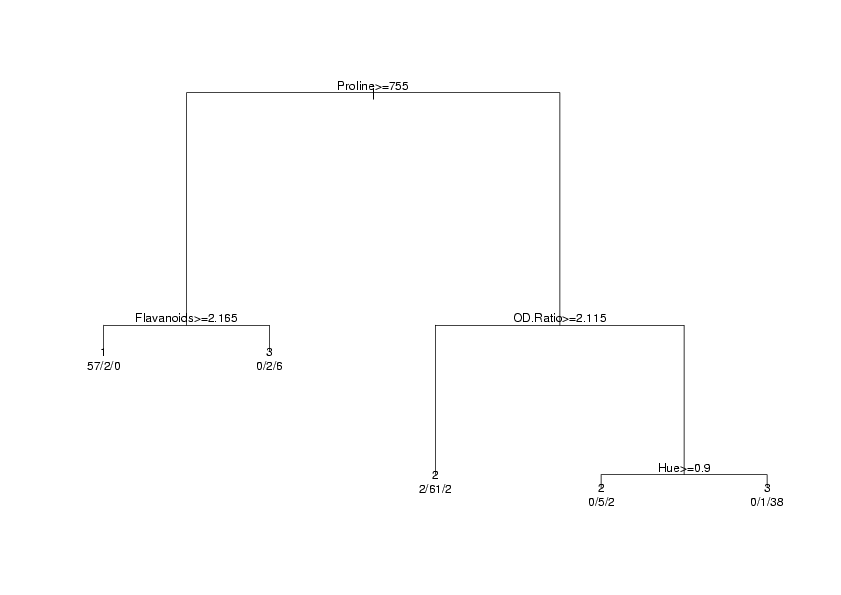 In order to calculate the error rate for an rpart analysis, we once again use the predict
function:
In order to calculate the error rate for an rpart analysis, we once again use the predict
function:
> pred.rpart = predict(wine.rpart,wine)
As usual we can check for names to see how to access the predicted values:
> names(pred.rpart)
NULL
Since there are no names, we'll examine the object directly:
> head(pred.rpart)
1 2 3
1 0.96610169 0.03389831 0.00000000
2 0.96610169 0.03389831 0.00000000
3 0.96610169 0.03389831 0.00000000
4 0.96610169 0.03389831 0.00000000
5 0.03076923 0.93846154 0.03076923
6 0.96610169 0.03389831 0.00000000
All that predict returns in this case is a matrix with estimated probabilities
for each cultivar
for each observation; we'll need to find which one is highest for each observation. Doing
it for one row is easy:
> which.max(rped.rpart[1,])
1
1
To repeat this for each row, we can pass which.max to apply, with a
second argument of 1 to indicate we want to apply the function to each row:
> table(apply(pred.rpart,1,which.max),wine$Cultivar)
1 2 3
1 57 2 0
2 2 66 4
3 0 3 44
To compare this to other methods, we can again use the row and col
functions:
> sum(tt[row(tt) != col(tt)]) / sum(tt)
[1] 0.06179775
Since rpart uses cross validation internally to build its decision rules, we can
probably trust the error rates implied by the table.
File translated from
TEX
by
TTH,
version 3.67.
On 19 Mar 2010, 15:13.
 It seems like the big differences are found in Flavanoids,
Hue and OD.Ratio
Now let's take a look at a method that makes it very clear which variables
are useful in distinguishing among the groups.
It seems like the big differences are found in Flavanoids,
Hue and OD.Ratio
Now let's take a look at a method that makes it very clear which variables
are useful in distinguishing among the groups.
 In order to calculate the error rate for an rpart analysis, we once again use the predict
function:
In order to calculate the error rate for an rpart analysis, we once again use the predict
function: