CGI Programming
1 A First CGI program with R
As a simple way of getting started with CGI programming, let's take the output
from the first simple form that we created and see how the information gets
translated from the CGI environment into the R environment.
I'm assuming that your SCF account
is s133xx and that you've followed the instructions at
http://www.stat.berkeley.edu/~spector/s133/projects/tech/Cgiprogs.html to
prepare your account to use CGI scripting. Typically, you would put the page
containing the html in your public_html directory. Suppose the
following is placed in a file called form1.html in your
public_html directory:
<html>
<form action='cgi-bin/R.cgi/test1.cgi' method=get>
<input type=text name=myvar><br>
<input type=submit value='GET'>
</form>
</html>
Since file names
in HTML are interpreted relative to the directory that the HTML resides in,
we can refer to the CGI program relative to the cgi-bin directory.
Notice that this will allow you to use the same form whether you're running on
the SCF network or through a tunnel.
In addition to
the formData list that holds the values of the CGI variables, information
is also passed to your CGI program through environmental variables. These
variables can be accessed with the Sys.getenv function, but for our
purposes here, we can use the showEnvironmentVariables function that's
part of the CGIwithR package. Here's a simple program that will just
print out some information about the formData list and the environmental
variables that are transfered into the environment:
Here's the test1.cgi program:
HTMLheader()
cat('Class of formData=',class(formData),' Mode of formData=',mode(formData),'<br>')
cat('Names of formData=',names(formData),'<br>')
tag(pre)
print(formData)
untag(pre)
showEnvironmentVariables()
cat('</body></html>')
If we run the program (by pointing a browser at http://springer/ s133xx/form1.html, entering "hello, world" and pressing the button), here's what the
output looks like:
Class of formData= list Mode of formData= list
Names of formData= myvar
$myvar
[1] "hello, world"
SERVER_SIGNATURE Apache/2.0.54 (Ubuntu) Server at springer Port 80
R_INCLUDE_DIR /usr/local/linux/R-2.2.1/include
HTTP_KEEP_ALIVE 300
HTTP_USER_AGENT Mozilla/5.0 (X11; U; Linux x86_64; en-US; rv:1.7.12) Gecko/20051010 Galeon/1.3.21 (Debian package 1.3.21-6ubuntu3) (Ubuntu package 1.0.7)
SERVER_PORT 80
HTTP_HOST springer
LD_LIBRARY_PATH /usr/local/linux/R-2.2.1/lib:/usr/local/lib64:/usr/lib/gcc/x86_64-linux-gnu/3.4.5:/usr/X11R6/lib:/server/linux/j2sdk1.4.2_04/jre/lib/i386/client:/server/linux/j2sdk1.4.2_04/jre/lib/i386
DOCUMENT_ROOT /mirror/data/pub/html/~spector/s133
HTTP_ACCEPT_CHARSET ISO-8859-1,utf-8;q=0.7,*;q=0.7
SCRIPT_FILENAME /class/u/s133/s133xx/public_html/cgi-bin/R.cgi
REQUEST_URI /~s133xx/cgi-bin/R.cgi/test1.cgi?myvar=hello%2C+world
SCRIPT_NAME /~s133xx/cgi-bin/R.cgi
R_GSCMD /usr/bin/gs
HTTP_CONNECTION keep-alive
PATH_INFO /test1.cgi
REMOTE_PORT 48242
PATH /usr/local/bin:/usr/bin:/bin
R_LIBS
PWD /class/u/s133/s133xx/public_html/cgi-bin
SERVER_ADMIN webmaster@localhost
R_SHARE_DIR /usr/local/linux/R-2.2.1/share
LANG
HTTP_ACCEPT_LANGUAGE en
PATH_TRANSLATED /class/u/s133/s133xx/public_html/cgi-bin/test1.cgi
HTTP_REFERER http://springer/~s133xx/form1.html
HTTP_ACCEPT text/xml,application/xml,application/xhtml+xml,text/html;q=0.9,text/plain;q=0.8,image/png,*/*;q=0.5
REMOTE_ADDR 128.32.135.22
SHLVL 1
SERVER_NAME springer
FORM_DATA myvar=hello%2C+world
SERVER_SOFTWARE Apache/2.0.54 (Ubuntu)
QUERY_STRING myvar=hello%2C+world
SERVER_ADDR 128.32.135.22
GATEWAY_INTERFACE CGI/1.1
R_HOME /usr/local/linux/R-2.2.1
SERVER_PROTOCOL HTTP/1.1
HTTP_ACCEPT_ENCODING gzip,deflate
R_DOC_DIR /usr/local/linux/R-2.2.1/doc
REQUEST_METHOD GET
R_SESSION_TMPDIR /tmp/RtmpPPrmxy
R_PLATFORM x86_64-unknown-linux-gnu
R_PAPERSIZE letter
R_PRINTCMD lpr
R_LATEXCMD /usr/bin/latex
R_DVIPSCMD /usr/bin/dvips
R_MAKEINDEXCMD /usr/bin/makeindex
R_RD4DVI ae
R_RD4PDF times,hyper
R_UNZIPCMD /usr/bin/unzip
R_ZIPCMD /usr/bin/zip
R_BROWSER /usr/bin/firefox
EDITOR vi
PAGER /usr/bin/less
R_PDFVIEWER /usr/local/linux/bin/acroread
AWK gawk
EGREP grep -E
MAKE make
PERL /usr/bin/perl
TAR tar
LN_S ln -s
R_USE_AQUA_SUBDIRS no
You may have noticed that the URL we used for the action=
specification makes it look like R.cgi is a directory, not
a file. What happens is that the R.cgi program gets
invoked by the web server, and it examines the PATH_INFO
variable to find the name of your script (test1.cgi in this case.)
It then loads up the formData list and calls your program.
If you examine the information that our simple web echoing program displayed,
you'll see that each of the headers that were passed to the program have
been transfered to environmental variables with a prefix of HTTP_.
Although you most likely won't need them, you can examine the list of
environmental variables to find information that you might want to use in
your CGI programs and then access the information by using Sys.getenv.
The most important thing to notice is that the myvar CGI variable,
defined in the input field of the HTML form, is available inside your R
program, in unencoded form, in the element with the name myvar. This
is the mechanism by which all the CGI variables are transfered into the R
environment.
2 Data
The R.cgi script calls your R program in such a way that it doesn't
automatically load any data into the R environment. So if you want to have
data available to your CGI program, you'll need to explicitly get the data
into R's environment.
For reasons of efficiency, your program should always use the load
function to load a previously saved binary version of the data you need.
The most convenient place to store these objects is right in the cgi-bin
directory from which your program will execute.
Suppose we wish to create a CGI program that will accept the name of one of the
variables from the wine data frame, and then display a summary of the
data. Before you plan to run the script, the wine data should be
saved in a simple R session that's started after you've changed your working
directory to be your cgi-bin directory. The command to do this is
save(wine,file='wine.rda')
Next, we can create a form, which would be saved in the public_html
directory. Here's a simple example, which we'll save in the file
wine.html:
<html><body>
<h1>Summary for Wine Variables</h1>
<form action='cgi-bin/R.cgi/dowine.cgi'>
Enter the name of the variable:
<input type=text name=winevar><br>
<center>
<input type=submit value='Run'>
</center>
</form>
</body></html>
The dowine.cgi program would look like this:
load('wine.rda')
HTMLheader()
winevar = formData$winevar
tag(h1)
cat('Summary for wine$',winevar,sep='')
untag(h1)
tag(h2)
tag(pre)
print(summary(wine[[winevar]]))
untag(pre)
untag(h2)
cat('</body></html>')
Here's the form:
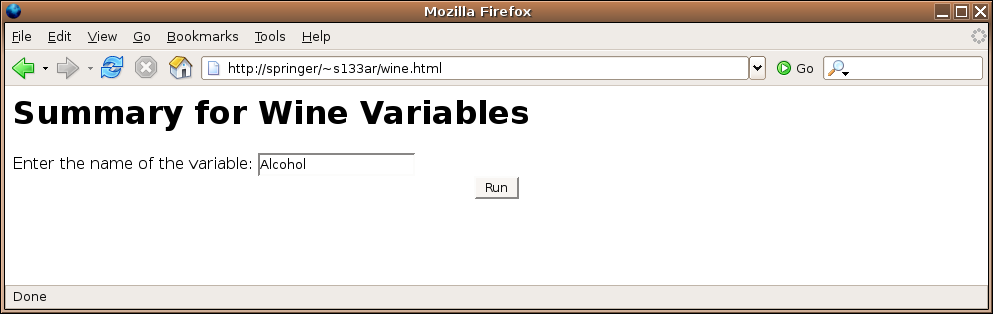 Here's the result of submitting the form:
Here's the result of submitting the form:
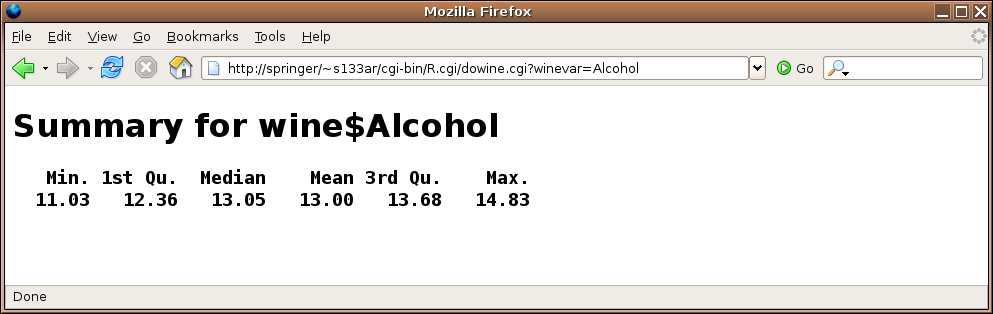
3 Combo Forms
Of course, having the user remember the name of the variable they're interested
in isn't a very user-friendly strategy, but the thought of manually preparing
a form that lists all the variables isn't very appealing either. The problem
can be solved by having the CGI program generate the form the first time it's
called, and then processing the form when it's submitted back to the web
server. If we call the CGI program directly (not through a form submission),
the formData list will be empty, and we can use that condition to
tell whether we need to generate the form or respond to it. Since R will be
generating the form, it's very easy to have it provide a choice for each variable.
For this example, let's use a drop down menu that will display the names of
each variable. Here's a program that will both generate the form and respond
to it:
if(length(formData) == 0){
HTMLheader()
tag(h1)
cat('Summary for Wine Variables')
untag(h1)
cat("<form action='dowine1.cgi'>")
cat("Choose the variable:")
cat("<select name='winevar'>")
load("wine.rda")
sapply(names(wine),function(x)cat("<option value='",x,"'>",x,"<br>\n",sep=''))
cat("</select>")
cat('<input type="submit" value="Run">')
cat("</form></body></html>")
} else {
load('wine.rda')
HTMLheader()
winevar = formData$winevar
tag(h1)
cat('Summary for wine$',winevar,sep='')
untag(h1)
tag(h2)
tag(pre)
print(summary(wine[[winevar]]))
untag(pre)
untag(h2)
untag(h2)
cat('</body></html>')
}
One very important thing to notice if you use this approach - the
action= argument should specify only the name of the program,
without the usual R.cgi/; since R is already calling your program,
it thinks it's in the R.cgi "directory".
Here's the result of calling the program directly:
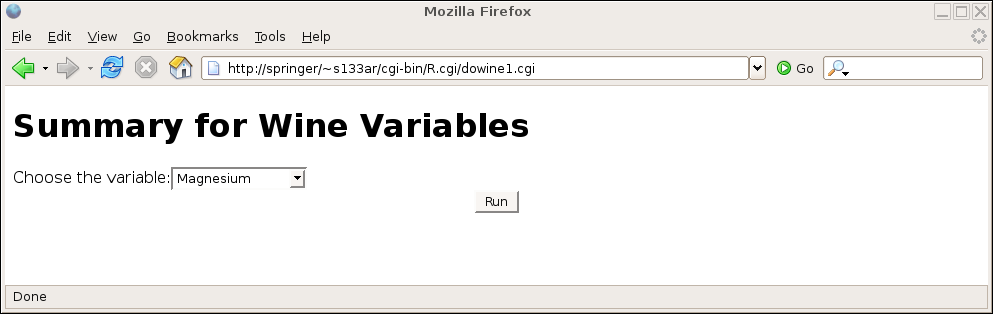 and here's the result after making a choice:
and here's the result after making a choice:
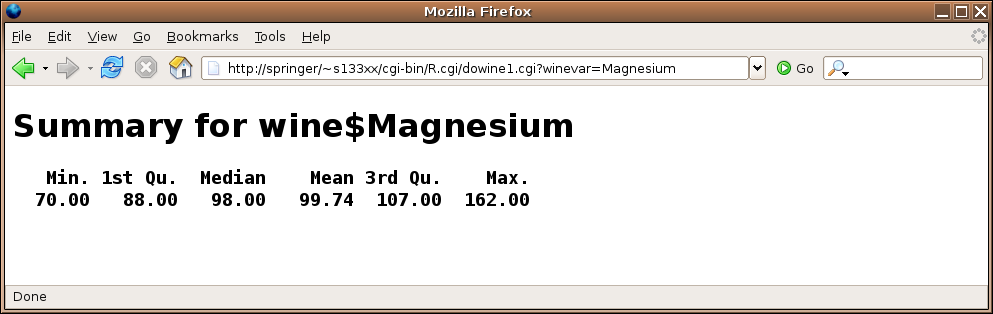
File translated from
TEX
by
TTH,
version 3.67.
On 17 Apr 2009, 15:49.
 Here's the result of submitting the form:
Here's the result of submitting the form:

 and here's the result after making a choice:
and here's the result after making a choice:
